 |

Instruments in the lab
The labs are located at two sites: Department of Pharmaceutical Sciences, on the south campus of University at Buffalo, and in the New York State Center of Excellence in Bioinformatics and Life Sciences (CoEBLS, the main lab) at downtown Buffalo, NY. These two sites encompass a total of 1200 sq ft of well-equipped wet-lab devoted to cell culture, biochemistry experiments (for Western-blotting, electrophoresis, ELISA, etc.), column packing, treatment of proteomic samples and preparation for quantitative analysis. Major equipment includes two ultra-centrifuges, one vacuum centrifuge, one lyophilizer, one set of solid phase purification system, one UV spectrometer, three hoods, one automatic cell counter and two incubators, as well as standard equipment for protein analysis.
There are two instrument rooms at 600 sq ft in total on the 3rd floor of CoEBLS, which house a number of state-of-the-art liquid chromatography (LC) systems and mass spectrometers:
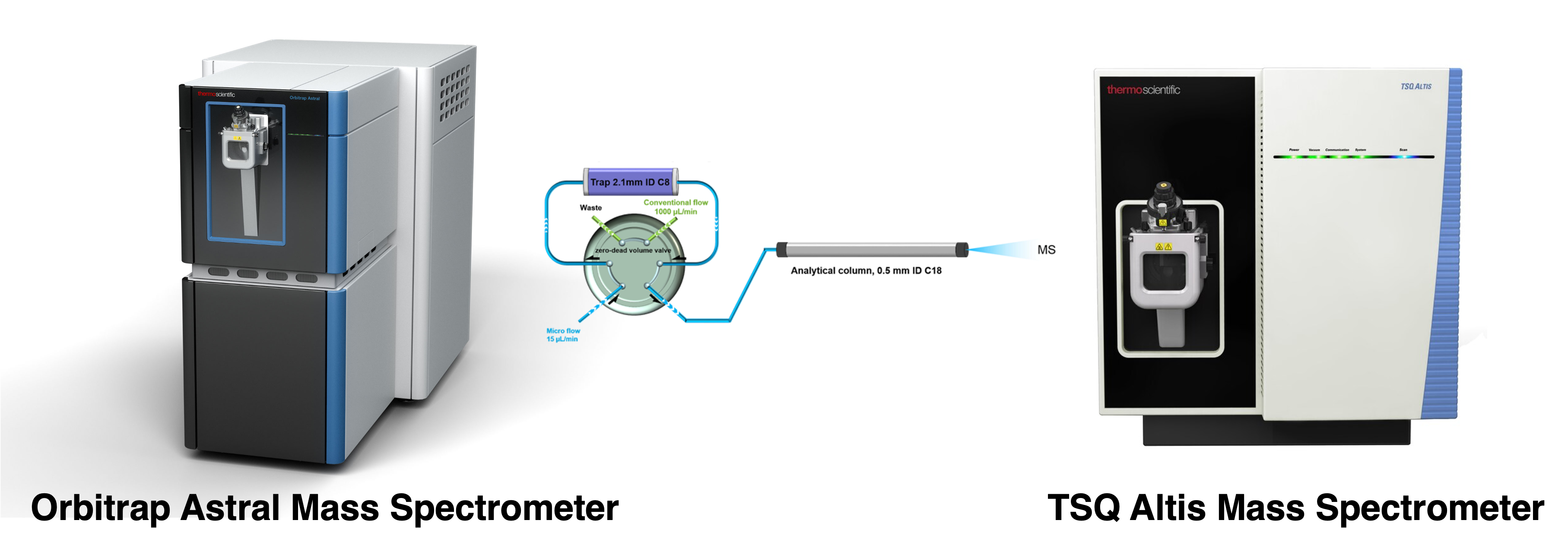
• Thermo Orbitrap Astral mass spectrometer
• Thermo TSQ Altis triple quadrupole mass spectrometer
• Thermo TSQ Quantiva triple quadrupole mass spectrometer
• Thermo Dionex Ultimate 3000 nano/micro HPLC systems with WPS-3000 autosamplers
• Waters Bioseparation semi-preparation HPLC system
• Several additional nano/micro LC systems (i.e. Eksigent nano-2D, Agient 1200-series micro HPLC, Shimadzu AC20 HPLC)

Among these instruments, the key ones are the three mass spectrometers in the lab. The Orbitrap Astral mass spectrometer can perform analyses with ultra-high mass resolution (>240,000) and mass accuracy (<3 ppm). The Astral mass analyzer provides unprecedented, deeper proteome coverage, higher sensitivity, and faster throughput than the previous mass analyzers. The Orbitrap Astral mass spectrometer sensitively quantifies more proteins in complex samples (e.g., neat plasma) and trace amounts of samples (e.g., single cells). The faster scan rate enables the use of shorter LC gradients and faster turnover time without compromising data quality. This revolutionary instrument provides in-depth and reliable proteomic identification/quantification of biological samples, enabling comprehensive characterization of molecular mechanisms underlying pathophysiological conditions, as well as the high-confidence discovery of putative biomarker candidates in patient plasma samples. It is also efficient for top-down proteomic analysis of low-molecular-weight proteins.
The TSQ instruments, on the othe rhand, offer ultra-high sensitivity and are able to significantly decrease chemical noises by using high-resolution selective reaction monitoring (SRM). Therefore, these instruments are employed for selective, sensitive and accurate quantification of specific target proteins in biological samples, e.g. antibodies biotherapeutics, antibody-drug conjugates, circulating biomarkers. The lab developed a trapping-micro LC-MS strategy to fulfill such needs, which achieves nano LC-MS-level sensitivity and conventional LC-MS-level throughput with extraordinary robustness.
UB Center for Computational Research
UB center for Computation Research (CCR) is located in the same building as the main laboratory. The CCR (www.ccr.buffalo.edu) is a leading academic supercomputing facility and maintains a high-performance computing environment and high-end visualization laboratories. Its technical staffs have broad expertise in scientific computing, numerical methods, software engineering, parallel processing, and visualization. The center's extensive computing facilities, which are housed in a state-of-the-art 4000 sq ft machine room, include a 2112 processor Intel Xeon-based (64-bit) Linux cluster, and a 64 processor shared memory Itanium-based cluster. CCR currently has more than 20 Tflops of aggregate computing capacity dedicated solely to support faculty led research projects at UB, and will soon undergo a $9M expansion to 50-70 Tflops of computing capacity (4000-6000 cores) and more than 800 Tbytes of high-performance storage capacity. CCR provides several layers of support for the life sciences/bioinformatics initiatives, including access to state-of-the-art hardware and software to facilitate research, the development of customized software/databases to support data analysis and workshops/training on software packages supported by CCR.
Currently the lab holds a 64-node concurrent license for SEQUEST, which is running on a Dell Linux cluster in the CCR. Both the above software and the hardware are dedicated to the lab. This will enable high-throughput analysis of the large amount of proteomic data that will be produced in this project.
Computer and software packages for data analysis

To extensively identify the myocardial proteins in swine plasma, the large amount of proteomic data will be analyzed both in the lab and in the UB CCR. In the lab, six high-end Dell work stations, as well as a high-performance data server are dedicated to data analysis. The primary software packages that we use and develop include: 1. Proteome Discoverer (Thermo Scientific) embedded with SEQUEST for protein identification; 2. TraceFinder (Thermo Scienfici) for analysis of targeted proteomics and metabolomics data; 3. SIEVE (Thermo Scientific), for chromatogram alignment and global recgonition of MS1 quantitative features; 4. PEAKS (Bioinformatics Solutions Inc.), for De Novo sequencing; 5. in-house developed R package, IonStarStat, for data incorporation, quantification and bioinformatics analysis. We own licenses for all of the above mentioned software packages.


