Issues
Divergence vs Convergence for similarity
Fitch (in Keim et al
(1981) until I find the better ref) proposed a test: consider:
with present day sequences at the top. Can test by adding more species to the tree
(dashed lines below for the newly added data. If we're dealing with divergence, then the
predicted ancestors (at the first nodes below) will be closer than the two initial
sequences; if however we're dealing with convergence, then the ancestors will be more
distant.
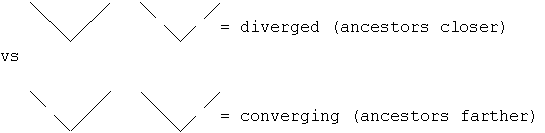
Good evidence for convergence is rare; best case I've seen is Stewart
& Wilson (1987). They found langurs have 10 amino acid substitutions in lysozyme
during the same period when baboons have only 4. More remarkably 5 of the 10 are shared
with the cow yet langurs are primates. They argue that this convergence is a response to
dietary selection with the langur eating leaves and having similar digestive constraints
to ruminants.
Duplication of genes and gene families and gene conversion
When look at many cloned genes, now obvious that within species there are great
similarities for different genes. ß globins in human are an example. If we have a cluster
of similar genes, then the cluster provides opportunities for further duplication and for
loss of genes by misalignment and crossing over. The resultant hybrid genes are more
similar to the ancestors than either ancestor is to each other. This process - gene
conversion - can occur even when the gene number is unaltered. The big issue is how to
detect it.
Need to do piecewise comparisons involving inter- and intra-specific alignments. An
example from our lab is that rats have multiple ß globin genes (probably 4) and mice have
2 (called ß-major and ß-minor). We have sequenced much of the genes of the rat and begun
the piecewise alignments.
Horizontal evolution and exchange of DNA segments
Stolzfuss
& Milkman (1988) examined the nature of genetic variation in DNA of populations of
E. coli. They find that clonal segments of DNA appear to be replaced not individual
bases. This makes good sense as a circular DNA requires two breakpoints for recombination.
It also appears that this mechanism may apply to natural populations of many bacteria.
More remarkable are similar observations for DNA exchange across conventional species
and genus lines in bacteria. Mazodier
& Davis (1988) have reviewed the observations to date. The data suggest that even
distantly related bacteria can exchange genetic material although the phenomenon is
normally detected only under strong selective pressure. The rate may be limited by the
lack of sequence similarity to drive efficient genetic recombination. In this sense all
bacteria can be thought of as one panmictic species!
The relation between phylogenetics and cladistitics
See Wheeliset
al (1992) for a microbiologically inspired analysis
.