Computational Approach to Modeling Microbiome Community Dynamics Associated with Chronic Human Disease Progression
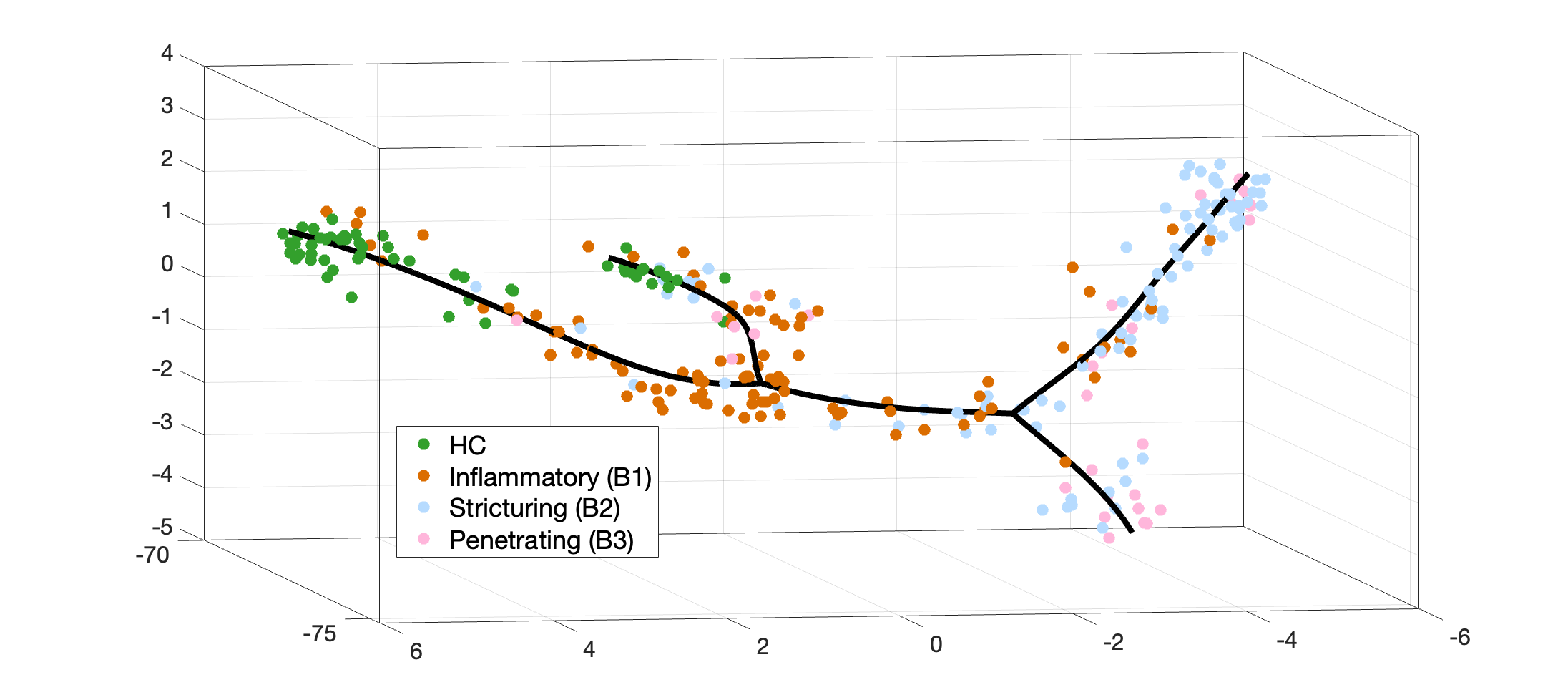
The human microbiome plays an essential role in many important physiological processes. However, due to the difficulty in obtaining longitudinal samples, the study of the dynamic relationship between the microbiome and human health remains a challenge. Here, we introduce a novel computational strategy that uses massive cross-sectional sample data to study microbial community dynamics associated with disease development. We tested the derived bioinformatics pipeline on a gut microbiome dataset from a Crohn's disease study. Our analysis revealed a double-bifurcating model that recapitulates the longitudinal progression of microbial dysbiosis observed during the clinical trajectory of the disease. By overcoming restrictions associated with complex longitudinal study sampling, the proposed strategy can provide valuable insights into the role of the microbiome in the pathogenesis of chronic disease and facilitate the shift of the field from descriptive research to mechanistic studies.
Documents
- Main paper
- Supplementary data
- Source code and documentation
