FDRnet: A Novel Method to Identify Significantly Perturbed Subnetworks in Cancer
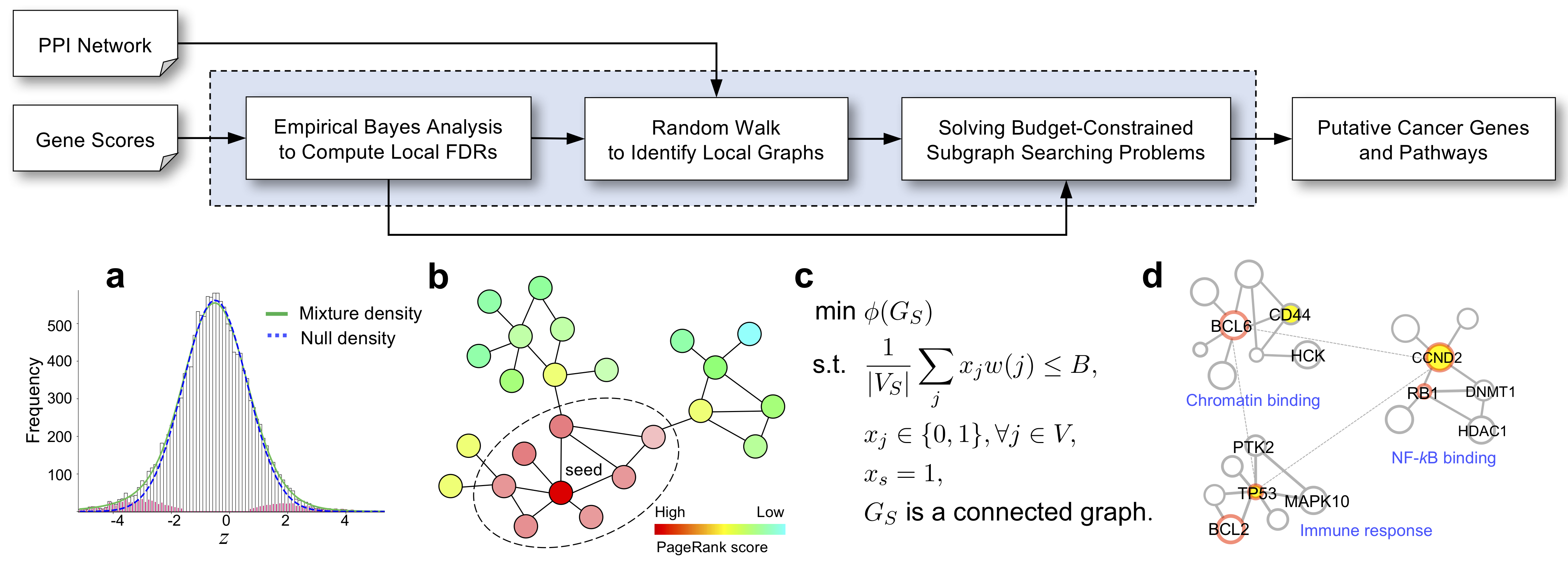
The identification of key functional networks using high-dimensional genomics data is critical for cancer research. Here, we introduce FDRnet, a novel method for the detection of significantly disrupted subnetworks in cancer, which addresses several key issues that have challenged the research community in pathway analysis. FDRnet detects significant subnetworks by solving a mixed-integer linear programming problem, using a given upper bound of false discovery rate (FDR) as a budget constraint and minimizing conductance scores to find dense subgraphs around seed genes. A large-scale benchmark study was performed on both simulation and cancer data. FDRnet demonstrated superior performance in terms of computational efficiency and the ability to detect functionally homogenous subnetworks in a scale-free biological network, to control FDRs of detected subnetworks, and to integrate multi-omics data. By overcoming the limitations of existing approaches, FDRnet can facilitate the detection of key functional pathways in cancer and other genetic diseases.
Documents
- Main paper
- Supplementary data
- Supplementary tables 8-11
- Software package and user manual
