|
|
|
Using Yeast Surface
Display to Engineer Protein
Deciphering the sequence-structure relationship in a polypeptide constitutes
an important goal in molecular biology. The large degrees of freedom available
even to a system of moderate size makes it difficult to arrive at a complete
description of the process by which the information required to fold to
a unique three-dimensional structure is encoded in a linear polypeptide
chain. One of the challenges impeding rapid progress in the field is the
difficulty of making detailed biophysical measurements on a large number
of protein variants to identify the underlying principles. This problem
is particularly poignant when studying proteins without a catalytic function
that can be easily and accurately measured. To study the sequence-structure
relationship in these proteins, one must resort to laborious and often slow
biochemical techniques. Even when the proteins that are being investigated
have known biological activities, e.g., affinity to a ligand or a function
critical to cellular survival, it is not always straightforward to decouple
the effects caused by structural changes from effects due to altered function.
Hence, they may equally require an independent biochemical analysis to confirm
results from a functional assay. Given the current available ensemble of
biochemical and biophysical techniques, it is clear that an efficienta highly
scalable experimental method of correlating sequence to structure and stability
would be an important development towards ultimately deriving predictive
rules that would to assist efforts to in the engineering of proteins for
with increased structural stability and consequent shelf-life in the case
of recombinant protein therapeutics.
Yeast surface display is a new display modality that's gaining popularity
in protein engineering (1-3). While somewhat new, yeast display is not much
different in its essence from other display technologies, including phage
display, ribosomal display and puromycin-based protein-DNA complexes. The
protein of interest is expressed on the yeast cell surface as fusion with
a mating factor protein Aga2p in a genetically modified yeast cell line
that stably expresses Aga1p. As Aga2p is expressed, it is directed to the
ER due to the signal sequence at the N-terminus, where it forms two disulfide
bonds with cell-surface bound Aga1p. From there, they are transported to
the cell wall together.
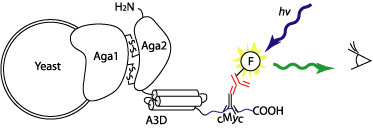 |
Figure 1. How yeast display works. The
level of fluorescence can be quantitatively measured by flow cytometry.
|
The expressed protein may be screened in a variety of ways. If the
protein has a function it may be directly assayed. For example, single chain
antibodies expressed on the yeast surface are fully functional and may be
screened based on binding to an antigen. Or if the protein doesn't have
any detectable function that can be easily assayed, its expression may be
monitored using an antibody. And because yeast is much larger than phage
one can use flow cytometry to monitor the phenotype of the protein on a
single yeast cell. Also yeast is an eukaryote, which means that sometimes
proteins that can't be well folded in prokaryotes such as E. coli may fold
well in yeast. We are using these unique properties of yeast surface library
to engineer stably folded proteins.
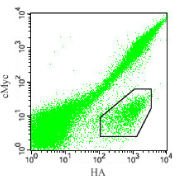 |
| Figure 2. The 2D cytogram of the a3D binary
peptide library. The cells were labeled with the c-Myc and HA antibodies.
A group of cells are labeled efficiently for the HA sequence but not
for the c-Myc sequence (marked with a polygon in the lower right corner).
|
Reference
1. Boder, E.T., and Wittrup, K.D. 1997. Yeast surface display for screening
combinatorial polypeptide libraries. Nat Biotechnol 15: 553-557.
2. Boder, E.T., and Wittrup, K.D. 1998. Optimal screening of surface-displayed
polypeptide libraries. Biotechnol Prog 14: 55-62.
3. Boder, E.T., and Wittrup, K.D. 2000. Yeast surface display for directed
evolution of protein expression, affinity, and stability. Methods Enzymol
328: 430-444.
|