|
|
|
Publications
34. Zhou, F.(*), Kroetsch, A.(*), Nguyen, V.P., Huang, X., Ogoke, O., Parashurama, N., Park, S.,
High affinity antibody detection with a bivalent circularized peptide containing antibody binding domains (Biotech J., accepted), (*) Equal contributions.
33. Kroetsch, A., Qiao, C., Heavey, M., Guo, L., Shah, D.K., Park, S., Engineered pH dependent recycling antibodies enhance elimination of Staphylococcal enterotoxin B superantigen in mice (mAbs, accepted) (doi: 10.1080/19420862.2018.1545510)
32. Kroetsch, A.(*), Chin, B.(*), Nguyen, V., Gao, J., Park, S. Functional expression of monomeric streptavidin and fusion proteins in Escherichia coli: applications in flow cytometry and ELISA, Applied Microbio & Biotech 102(23), 10079-10089 (2018), (*) Equal contributions.
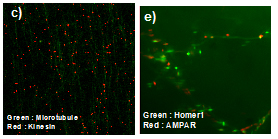
31. S. H. Lee, C. Jin, E. Cai, P. Ge, Y. Ishitsuka, K. W. Teng, A. A. de Thomaz, D. Nall, M. Baday, O. Jeyifous, D. Demonte, C. M. Dundas, S. Park, Delgado, J., W. N. Green, P. R. Selvin. Super-resolution Imaging of Synaptic and Extrasynaptic Pools of Glutamate Receptors with Different-sized Fluorescent Probes, eLife e27744 (2017) (doi: 10.7554/eLife.27744)
30. Mann, J.K., Shen, J., Park, S. Enhancement of muramyl dipeptide-dependent NOD2 activity by a self-derived peptide J Cell Biochem 118:1227-1238 (2017)
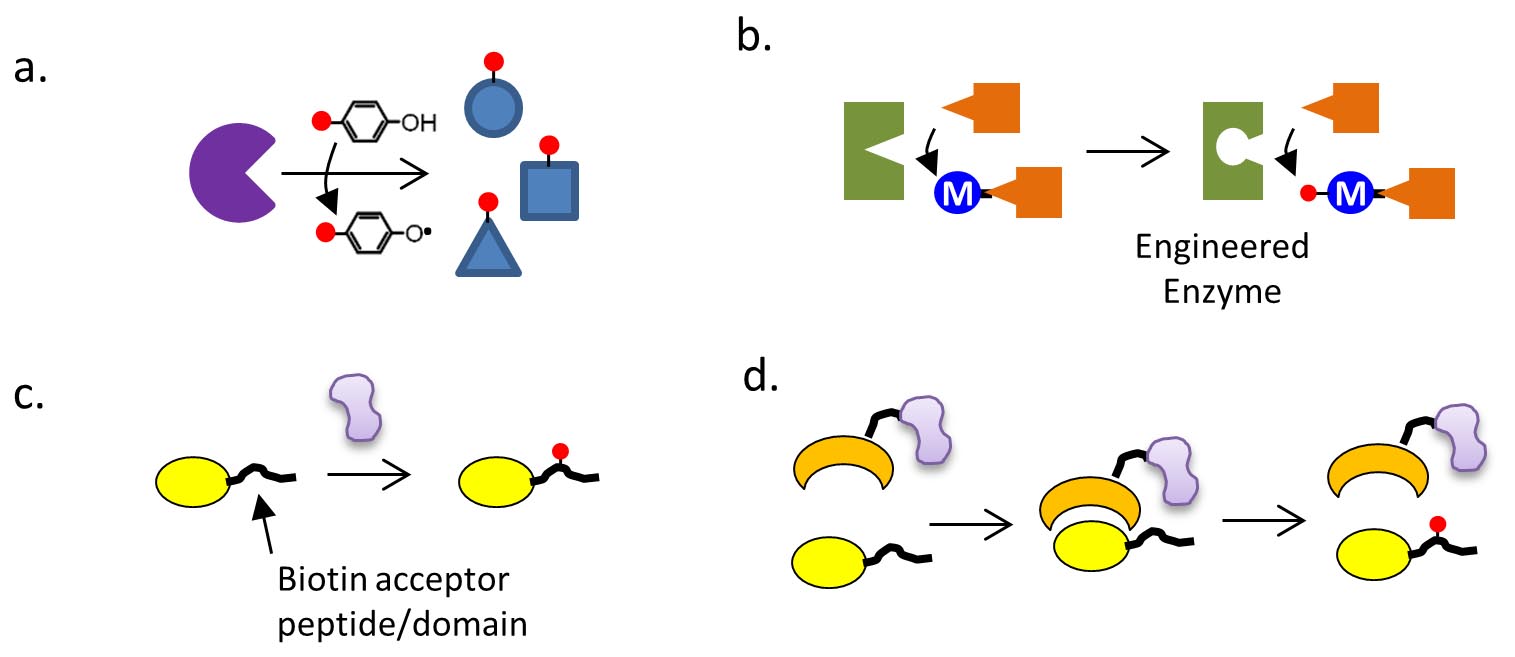
29. Mann, J.K.(*), Demonte, D.(*), Dundas, C.M., Park, S., Cell labeling and proximity dependent biotinylation with engineered monomeric streptavidin, Technology 4, 1-7 (2016), DOI: 10.1142/S2339547816400057
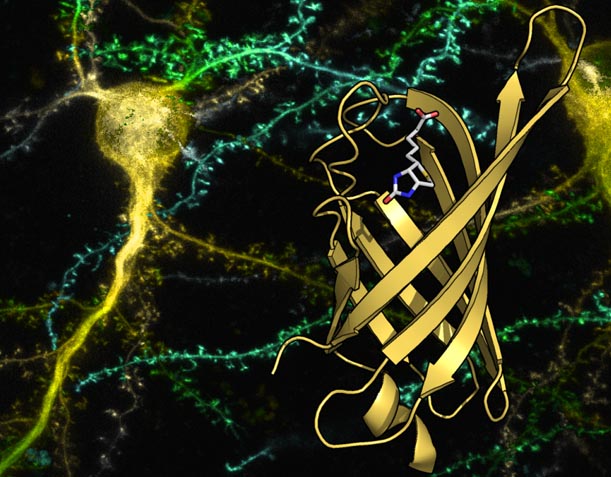
28. Chamma, I., Letellier, M., Butler, C., Lim, K.H., Gauthereau, I. Choquet, D., Sibarita, J.B., Park, S., Sainlos, M., Thoumine, O., Mapping the dynamics and nanoscale organization of synaptic adhesion proteins using monomeric streptavidin, Nature Commun 7:10773 (2016) (DOI: 10.1038/ncomms 10773)
27. Demonte, D., Li, N., Park, S., Postsynthetic domain assembly with NpuDnaE and SspDnaB split inteins. App Biochem Biotech 177, 1137-51 (2015)
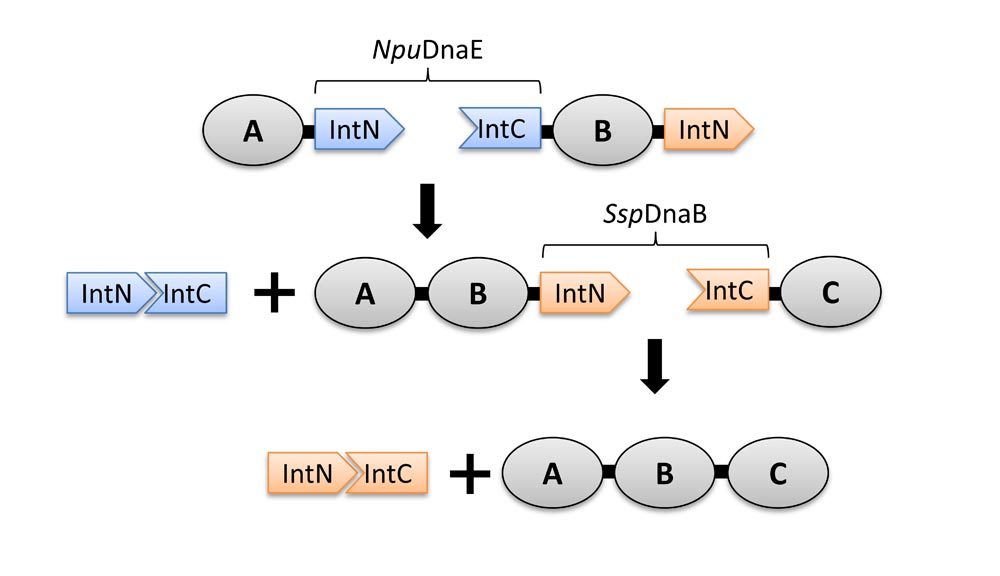
26. Kroetsch, A. and Park, S. More than one way to skin a cat: in-situ engineering of an antibody through photo-conjugated C2 domain, Biotechnol J 10, 508-509 (2015)
25. Wang, H., Carrier, S., Park, S., Schultz, Z.D., Selective TERS Detection and Imaging through Controlled Plasmonics, Faraday Discussions 178:221-35 (2015)
24. Demonte, D., Dundas, C.M., Park, S., Expression and purification of soluble monomeric streptavidin in E. coli. App Microbiol & Biotech 98, 6285-6295 (2014)
23. Mann, J.K., Park, S. Epitope specific binder design by yeast surface display. Meth Mol Biol 1319:143-54 (2015)
22. Dundas, C.M., Demonte, D., Park, S., Streptavidin-biotin technology: improvements and innovations in chemical and biological applications. App Microbiol & Biotech
97:9343-9353 (2013)
21. Park, S., Mann, J.K., Li, N., Targeted inhibitor design: lessons from small molecule drug design, directed evolution, and vaccine research. J SciMed Central CEPT 1:1004(2013). (read).
20.
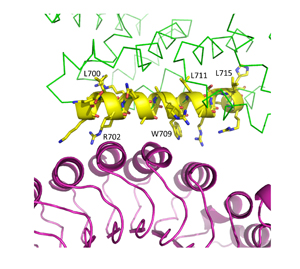
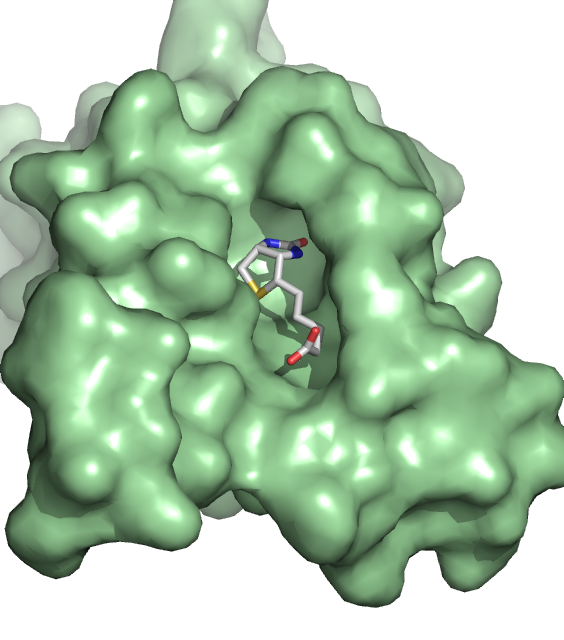
Demonte, D., Drake, E., Lim, K.H., Gulick, A., Park, S., Structure based engineering of streptavidin monomer with a reduced biotin dissociation rate. Proteins: Structure, Function, and Bioinformatics 81, 1621-1633 (2013)
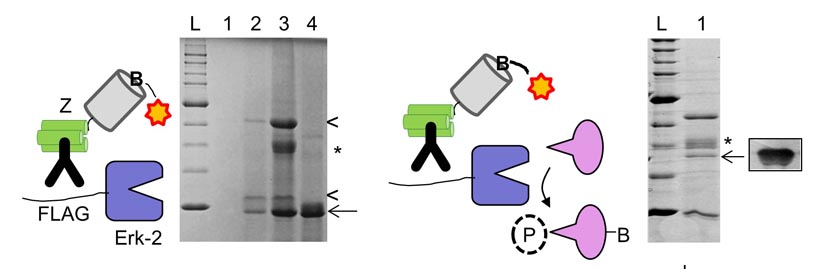
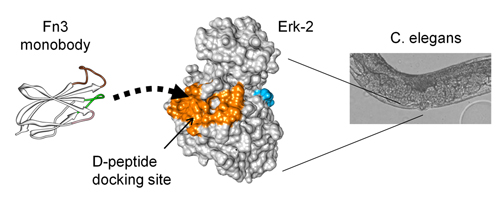
19. Mann, J.K., Wood, J.F., Stephan, A.F., Tzanakakis, E.S., Ferkey, D.M., Park, S. Epitope guided engineering of monobody binders for in vivo inhibition of Erk-2 signaling. ACS Chem Biol 8, 608-616 (2013)
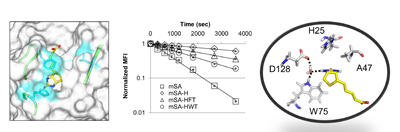
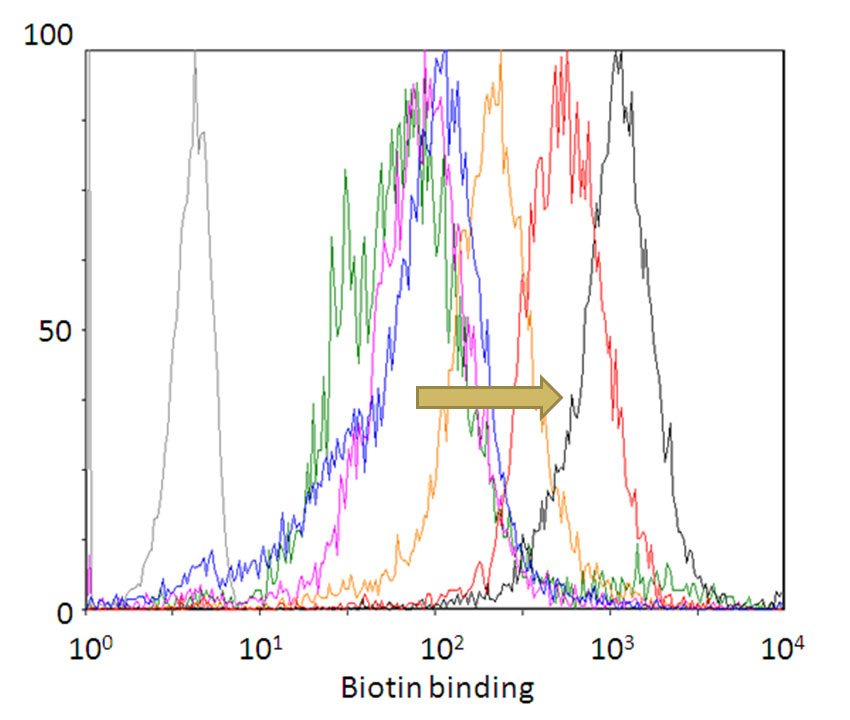
18. Lim, K.H., Huang, H., Pralle, A., and Park, S. Stable, High-Affinity Streptavidin Monomer for Protein Labeling and Monovalent Biotin Detection. Biotechnology Bioeng 110, 57-67 (2013)
(Video abstract available on youtube.)
(Plasmids available at Addgene: pRSET-mSA, pRSET-mSA-EGFP, pDisplay-mSA-EGFP-TM, pYD1-mSA.)
17. Lim, K.H., Huang, H., Pralle, A., and Park, S. Engineered streptavidin monomer and dimer with improved stability and function. Biochemistry 50, 8682-8691 (2011)
16. Lim, K.H., Hwang, I. and Park, S.
Biotin-assisted folding of streptavidin on the yeast surface. Biotechnol Prog 28, 276-283 (2011)
15. Hsu, C.K. and Park, S. Computational and mutagenesis studies of the streptavidin native dimer interface. J Mol Graph Model 29, 295-308 (2010)
-Featured as "Topical Perspectives"
14. Lim, K.H., Hsu, C.K., Park, S. Flow cytometric analysis of genetic FRET detectors containing variable substrate sequences. Biotechnol Progress 26, 1765-1771 (2010)
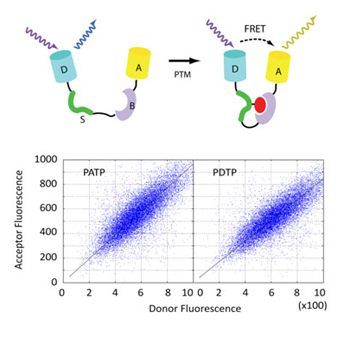
13. Lim, K.H., Madabhushi, S.R. (*), Mann, J. (*), Neelamegham, S., Park, S. Disulfide trapping of protein complexes on the yeast surface. Biotechnol Bioeng 106, 27-41 (2010). *Equal contributions.
- Featured in Spotlight ("Getting STUCKED")
12. Sheldon Park and Jennifer Cochran (Eds). Protein Engineering and Design, CRC Press (2009)

11. Hwang, I. and Park, S. Computational design of protein therapeutics. Drug Disc Today: Tech 5, e43-48 (2008).
10. Szep,S.(*), Park, S.(*#), Boder, E.
T., Van Duyne, G., Saven, J. G. Structural coupling between FKBP12 and buried water. Proteins: Structure, Function, and Bioinformatics 74, 603-11 (2009). (*) Equal contributions, (#) Corresponding author.
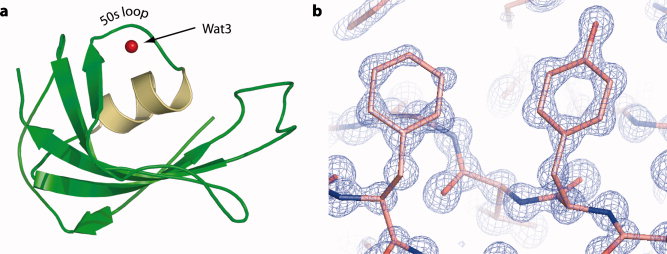
9. Park, S., Xu, Y., Stowell, X. F., Gai, F., Saven, J. G., Boder,
E. T. Limitatioins of yeast surface display
in engineering proteins of high thermostability. Protein
Eng Des Sel (PEDS) 19, 211-217 (2006).
8. Park, S. & Saven, J. G. Simulation of pH-dependent
edge strand rearrangement in human b-2 microglobulin. Protein
Sci 15, 200-207 (2006).
7. Park, S. & Saven, J. G. Statistical and
molecular dynamics studies of buried waters in globular proteins. Proteins 60, 450-463 (2005).
6. Park, S., Saven, J. G. Computationally assisted
protein design. Annu Rep in Compu Chem, Volume 1,
Chapter 18, pp245- 253 (2005).
5. Park, S., Boder, E. T., Saven, J. G. Modulating
the DNA affinity of Elk-1 with computationally selected mutations. J Mol Biol 348, 75-83 (2005).
4. Park, S., Kono, H., Wang, W., Boder, E. T.,
Saven, J. G. Progress in the development and application of
computational methods for probabilistic protein design. Comp
Chem Eng 29, 407 (2005).
3. Park, S., Fu, X., Wang, W., Yang, X., Saven,
J. G. Computational protein design and discovery. Annu
Rep Prog Chem Sect C 100, 195-236 (2004).
2. Park, S., Yang, X., Saven, J. G. Advances in
computational protein design. Curr Opin Struct Biol 14, 487-494 (2004).
1. Park, S., Uesugi, M., Verdine, G. L. A second
calcineurin binding site on NFAT regulatory domain. Proc
Natl Acad Sci U S A 97, 7130-5 (2000).
Patent
Sheldon Park and Kok Hong Lim, Compositions Comprising Monomeric Streptavidin and Methods for Using Same, US Patent filed (Application no. PCT/US10/32542)
|