Advanced Linux
BCH 519
Spring 2021
Video Recordings
Videos for BCH519 Unit 2 can be found at:
Outline for this lecture
- Recapping Linux Basics
- A Few Additional Commands
- Working Through Some Examples
- Gene Expression and File Reformatting
Text Manipulation
- Search for patterns in file:
grep - Find differences between files:
diff - Print first/last n lines:
head, tail - Print selected parts of file:
cut - Sort file:
sort
$ grep "pattern" my_file
$ diff -u file1 file2
$ head -n 20 my_file
$ tail -n 20 my_file
$ cut -f 1,3 my_file
$ sort my_file
Input/Output redirection
>takes output from command and writes it to file>>takes output from command and appends it to file<takes input from file and sends it to command
$ echo "Hello World" > hello.txt
$ cat hello.txt
Hello World
$ echo "Goodbye" >> hello.txt
$ cat < hello.txt
Hello World
Goodbye
$ grep "Hello" hello.txt > results.txt
$ cut -f 1,3 output.txt > columns-1-3.txt
Pipes
- Allows you to “chain” commands together
|= pipe|take output from command on left and send as intput to command on right
$ cat my_file.txt | wc -l
$ cat my_file.txt | grep -i "hello"
$ echo "Hello World" | wc -c > num-chars.txt
$ echo "1234" | rev
4321
Find
- Find . . . finds files :)
- find . -maxdepth 1 -type d -mtime +365
# Find all files
$ find .
# Find all directories
$ find . -type d
# Easily find all folders that are older than 365 days
$ find . -type d -mtime +365
sed
- Stream editor
sed 's/find/replace/g'- Replaces one or more occurances of a pattern
- Similiar to Find and Replace in MS Office
- Example: Renaming chrIV to chr04
$ echo "foobar"
foobar
$ echo "foobar" | sed 's/foo/bar/'
barbar
Using paste
- Quick way to join two files side-by-side
-d: specify delimiter (-d "\t")
$ head -n 1 blacklist-names.txt
chr1 564449 570371 High_Mappability_island
$ head -n 1 blacklist-score.txt
chr1 564449 570371 1000 +
$ paste -d"\t" blacklist-names.txt blacklist-score.txt | head -n 1
chr1 564449 570371 High_Mappability_island chr1 564449 570371 1000 +
Using paste continued..
- Multiple commands with redirection are very powerful
- This string of commands takes two files, pastes them together, and reformats it with cut into a properly formatted BED file
$ paste -d"\t" blacklist-names.txt blacklist-score.txt | head -n 1
chr1 564449 570371 High_Mappability_island chr1 564449 570371 1000 +
$ paste -d"\t" blacklist-names.txt blacklist-score.txt | cut -f1,2,3,4,8,9 | head -n1
chr1 564449 570371 High_Mappability_island 1000 +
AWK: Versatile Scripting
- Initially developed in 1977 by Alfred Aho, Peter Weinberger, and Brian Kernighan
- Extremely versatile stream editing tool
- Column Order, Arthimetic, Data Transformations
awk condition {action}
AWK: Example
- More flexible than cut, but steeper learning curve
- Often the last step to finalize column order
$ awk '{print $1,$2}' < blacklist-names.txt | head -n 1
chr1 564449
$ awk '{print $4,$2,$1}' < blacklist-names.txt | head -n 1
High_Mappability_island 564449 chr1
AWK: Sample Scripts
- Capable of doing some pretty awesome things
# Print the number of fields in each line, followed by the line
$ awk '{ print "Field Count:" NF "\t" $0 } ' fimo.bed | head -n1
Line Count:6 chr2 232477432 232477450 MA0139.1 27.1967 +
# Print fields in reverse order
$ awk '{for (i=NF; i>0; i--) printf("%s ",$i);print ""}' fimo.bed | head -n 1
+ 27.1967 MA0139.1 232477450 232477432 chr2
Examples using real data
- Processing tabbed gene-expression data
- Producing an expression table for post-processing
- Common file reformat (FIMO to BED)
Parsing Cufflinks Gene Expression
Cufflinks is an industry standard tool used to compute per-gene expression levels directly from Next-Generation sequencing data. It produces a tabular format file, one table per sample.$ ls *.cufflinks.txt
5952_SRR015086.cufflinks.txt 5952_SRR015088.cufflinks.txt
5952_SRR015090.cufflinks.txt 5952_SRR015092.cufflinks.txt
$ head -n 2 5952_SRR015086.cufflinks.txt
tracking_id class_code nearest_ref_id gene_id gene_short_name tss_id locus length coverage FPKM FPKM_conf_lo FPKM_conf_hi FPKM_status
FBgn0085737 - - FBgn0085737 CR40502 - 211000022278158:591-1036 - - 212.084 184.737 239.432 OK
Cutting cufflinks tables
- Interested in gene_id, locus, and FPKM value
- Columns 4, 7 and 10 respectively
cut -f4,7,10 *.cufflinks.txt- Store output in temp files
$ for i in *.cufflinks.txt; do cut -f4,10 $i > $i.temp ; done
$ head -n 2 *.temp
==> 5952_SRR015086.cufflinks.txt.temp <==
gene_id FPKM
FBgn0085737 212.084
==> 5952_SRR015088.cufflinks.txt.temp <==
gene_id FPKM
FBgn0085737 0
Sorting the temp files
- We need to maintain gene order!!
sort -k1,1 -k2,2
$ for i in *.temp; do sort -k1,1 -k2,2 $i > $i.sorted ; done
$ ls *.sorted
5952_SRR015086.cufflinks.txt.temp.sorted 5952_SRR015090.cufflinks.txt.temp.sorted
5952_SRR015088.cufflinks.txt.temp.sorted 5952_SRR015092.cufflinks.txt.temp.sorted
Pasting cufflinks temp files
- Create a multi-column file for all samples
paste -d"\t":-dspecifies tab delimited- Note: this will paste the files side in order of name
$ paste -d"\t" *.sorted | cut -f1,2,3,6,9,12 > Gene-Table.txt
$ tail Gene-Table.txt
FBtr0347422 Y:3272834-3273532 0 0 0 0
FBtr0347466 Y:3267087-3268196 13.4179 0 15.302 13.4467
FBtr0347566 Y:3047605-3048886 7.18664 0 7.04498 7.88644
FBtr0347604 3L:21949824-21951550 0 0 0.0992518 0.195406
FBtr0445173 X:20217401-20218244 16.3069 0 17.2985 18.7958
FBtr0445175 X:20209745-20210592 20.2268 0 19.9883 23.4011
FBtr0445177 X:20202075-20202922 20.4815 0 19.9792 23.2494
FBtr0445213 X:8397156-8398298 1.40773 0 1.81782 2.47734
FBtr0445214 X:1163619-1164105 0 0 0 0
Viewing the Table
- Any text editor or spreadsheet application will do!
Reformatting Files using Linux
- A very common request . . .
- “Can you reformat from X to Y?”
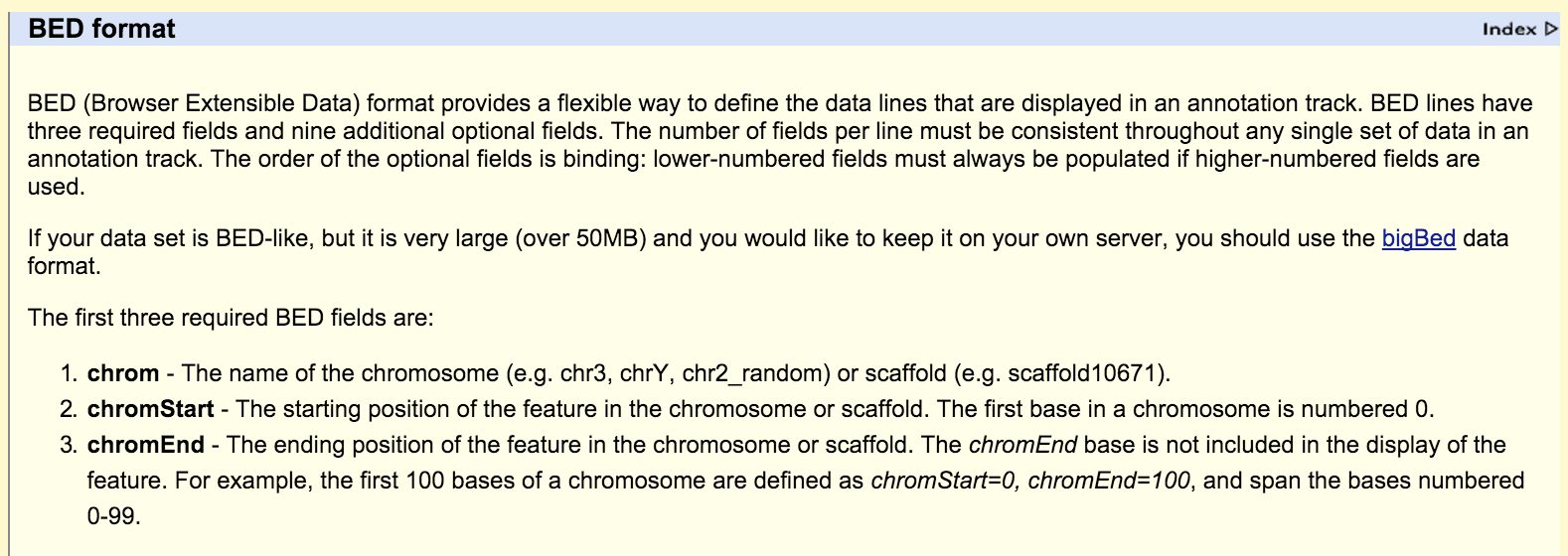
UCSC File Format Resource
https://genome.ucsc.edu/FAQ/FAQformat.html
FIMO to BED Format
FIMO is a command line tool that is part of the MEME-Chip analysis package. It is capable of searching a DNA sequence for matches to transcription factor motifs. Unfortunately, the output is not compatible with most downstream analysis tools. It's output is the following:| FIMO Format | BED Format |
|---|---|
|
|
Format Exercise: Fimo to Bed
$ more fimo.txt
#pattern name sequence name start stop strand score p-value q-value matched sequence
MA0139.1 chr2 232477432 232477450 + 27.1967 4.53e-12 0.00752 TGGCCACCAGGGGGCGCCG
MA0139.1 chr16 85004316 85004334 - 27.1311 6.58e-12 0.00752 CGGCCACCAGGGGGCGCCA
MA0139.1 chr3 97742256 97742274 - 27.1311 6.58e-12 0.00752 CGGCCACCAGGGGGCGCCA
MA0139.1 chr5 136835086 136835104 - 27.1311 6.58e-12 0.00752 CGGCCACCAGGGGGCGCCA
$ awk '{print $2,$3,$4,$1,$6,$5}' fimo.txt
name sequence name #pattern stop start
chr2 232477432 232477450 MA0139.1 27.1967 +
chr16 85004316 85004334 MA0139.1 27.1311 -
chr3 97742256 97742274 MA0139.1 27.1311 -
chr5 136835086 136835104 MA0139.1 27.1311 -
Format Exercise: Fimo to Bed
Pipe output into `grep -v` to invert match and remove header line$ awk '{print $2,$3,$4,$1,$6,$5}' OFS="\t" < fimo.txt | grep -v name > fimo.bed
chr2 232477432 232477450 MA0139.1 27.1967 +
chr16 85004316 85004334 MA0139.1 27.1311 -
chr3 97742256 97742274 MA0139.1 27.1311 -
chr5 136835086 136835104 MA0139.1 27.1311 -
Sorting Data
- Sorting BED format is standard practice
- Allow for indexing, increasing speed of access
-k, --key=POS1[,POS2]start a key at POS1, end it at POS2 (default end of line)
$ sort -k1,1 -k2,2n -k3,3n
chr16 85004316 85004334 MA0139.1 27.1311 -
chr2 232477432 232477450 MA0139.1 27.1967 +
chr3 97742256 97742274 MA0139.1 27.1311 -
chr5 136835086 136835104 MA0139.1 27.1311 -
In-Class Exercises
- Use cut to view the first three colums from blacklist-names.txt
- Use awk to view the first three colums from blacklist-names.txt
- Paste the blacklist-names.txt and blacklist-score.txt files, and use cut or awk to generate a properly formatted bed file
