
 |
Computational Biophysics Group |  |
| Research | Publications | Open Positions | ENM webserver |
| Welcome to the Computational Biophysics Group of University at Buffalo! We currently have openings for graduate research assistant and postdoc. |
|
Research Conformational dynamics or motions hold keys to the biological functions carried out by biomolecules such as proteins and nucleic acids. Biologically relevant motions, spanning a wide range of time scales (ns ~ min), are often prohibitively expensive to simulate by standard computational techniques like molecular dynamics which is based on atomistic force fields. Alternatively, highly simplified coarse-grained models promise to efficiently probe biomolecular dynamics of large systems and long time scales. For example, the elastic network model (ENM), explored in Dr.Zheng's recent work (see publications), represents a protein as a 3D network of beads connected by harmonic springs (Fig 1). The normal mode analysis of ENM yields a spectrum of normal modes that capture various protein conformational changes (such as inter-domain hinge motions) observed in structural biology. The computational tools that I have developed based on ENM are now availabe in a webserver. Recently, new computational methods for modeling transition pathways, flexibly fitting cryo-electron-microscopy maps and solution X-ray scattering data have been developed and added to our webserver. For details, please check out our publications. Our long-term goal is to investigate biomolecular functions through the multi-scale modeling of biomolecular dynamics at a wide range of length scales from whole molecules down to individual atoms. We will employ various structural models of biomolecules from coarse-grained to atomistic models. These complementary models will be welded coherently to achieve both efficiency and accuracy. We are interested in integrating computer modeling with experiments via collaborations so that our models are correctly parameterized by fitting experimental data, and then make informative predictions to guide future experiments. Our ultimate goal is to computationally elucidate the molecular functions of various molecular motors such as myosins and kinesins . |
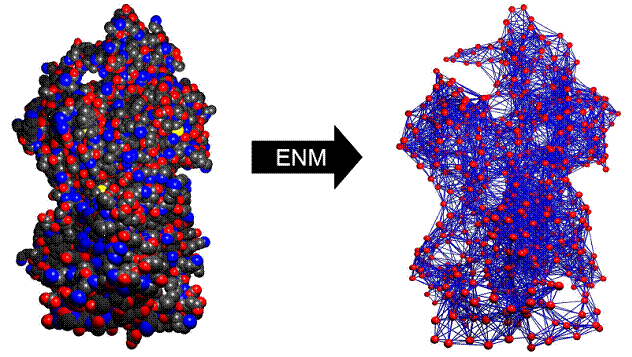
Fig 1. Building an elastic network model (right) from an atomistic crystal structure (left) |
|
Gallery | |||
 |
 |
 |
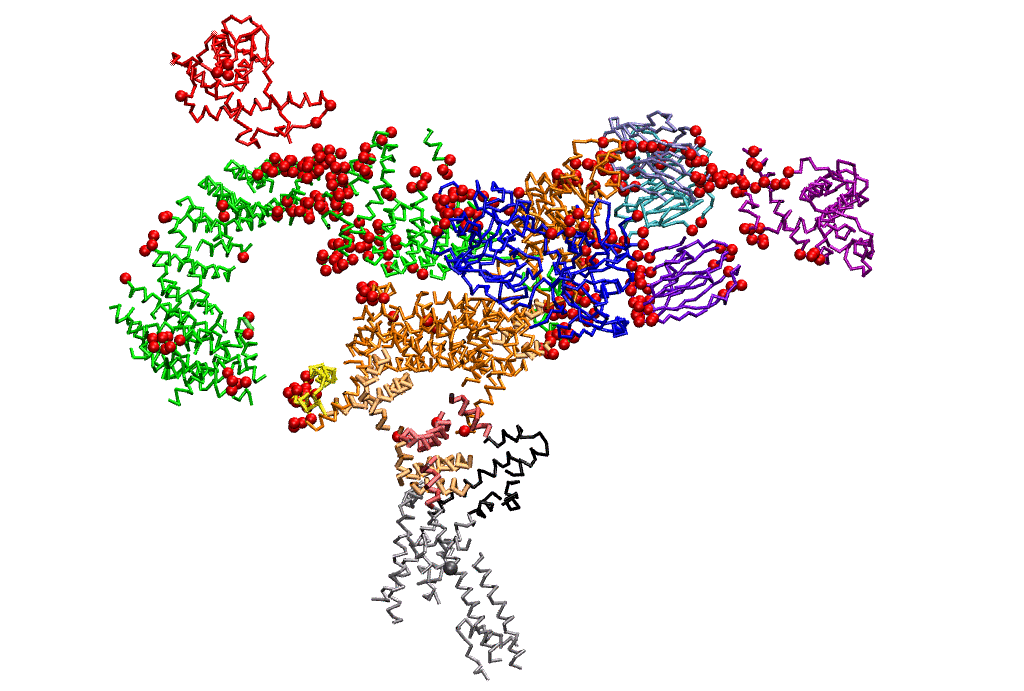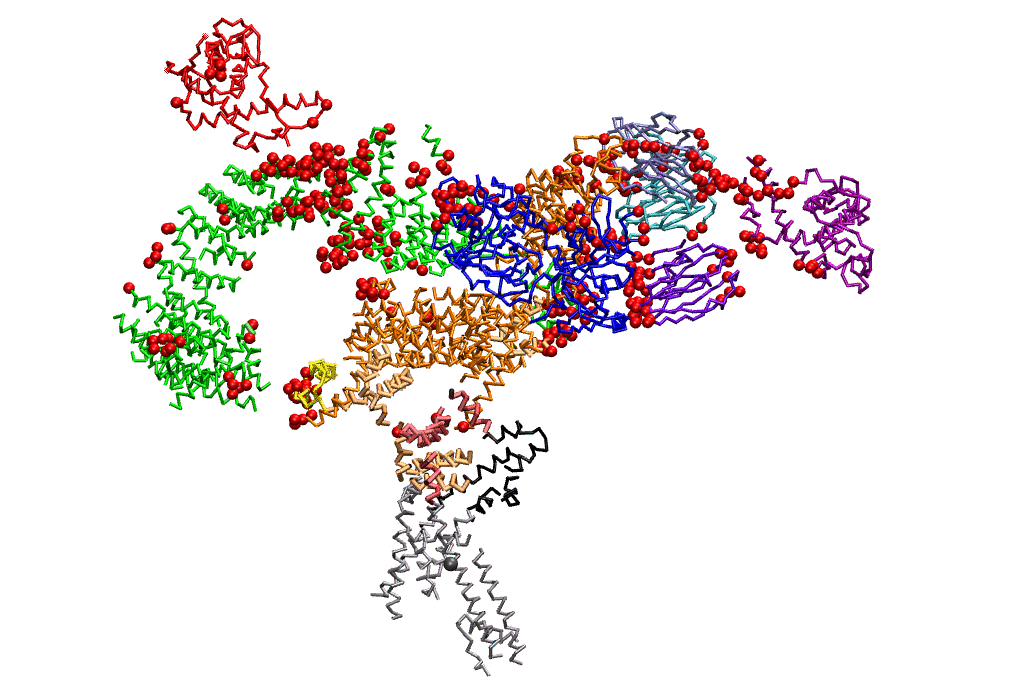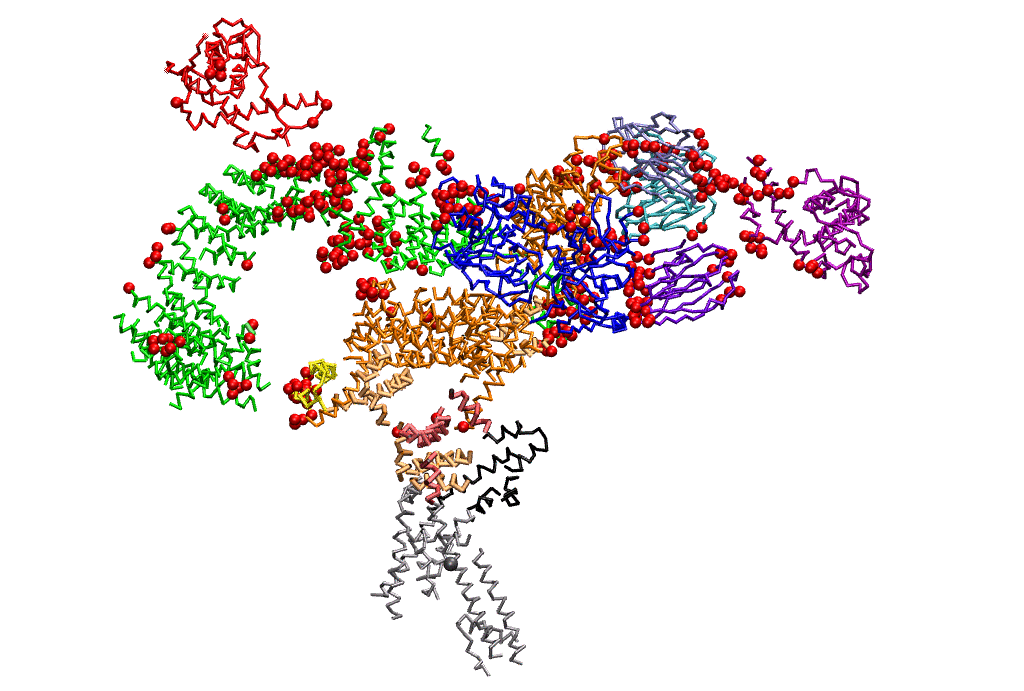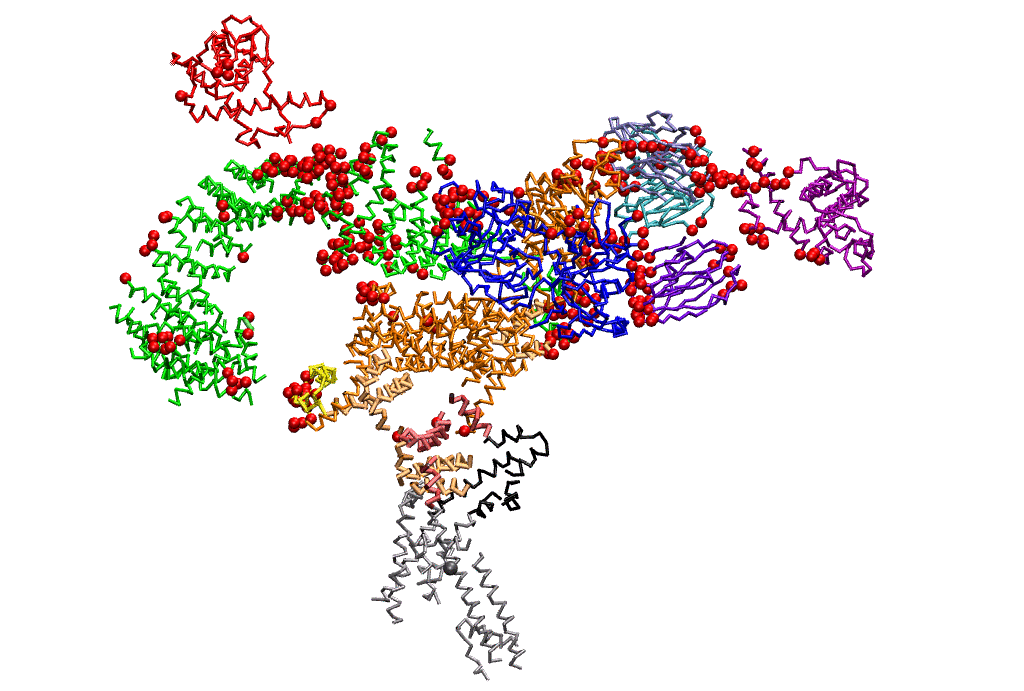 |
| Dynein's power stroke | NS3 helicase translocating DNA | Mode 6 of TRPV1 (PDB) | Mode 2 of Ryanodine receptor (PDB) |
Last update 09/01/2012