 Gerald B. Koudelka
Gerald B. Koudelka
Professor of Biological
Sciences
College of Arts &
Sciences
DNA-Protein Interactions; DNA Structure,
Transcriptional Regulation
Bacterial Pathogenesis
Ph.D. 1984 University at Buffalo
Postdoctoral work 1984-88
Harvard University
Address Information
Gerald B.
Koudelka
Department of Biological Sciences
607 Cooke Hall
State University of New York at Buffalo
Buffalo, NY 14260
Phone: (716) 645-4940
(Research Office)
Click for Email
RESEARCH SUMMARY
The research in
the Koudelka lab is focused around two central themes:
Mechanisms of Indirect Readout:
The mechanisms
whereby regulatory proteins recognize specific DNA sequences remains one of the
most important areas of study in biology. This process requires that the
protein be able to seek out and recognize its particular
binding sequence, in the presence of an overwhelming number of potential
non-specific binding sites. In our studies of direct readout of DNA sequence,
we have uncovered the intimate details of how amino acids and base pairs can
interact, and how these interactions can be regulated by both protein and DNA
structure. In indirect readout,
sequence-dependent differences in the structure and flexibility of noncontacted
bases in a DNA binding site regulate the stability and sequence-specificity of
a protein-DNA complex. Despite the high prevalence and functional importance of
indirect readout it is unclear how DNA sequence differences lead to changes in
DNA structure and flexibility. We are determining the structural basis for, and
functional implications of the indirect readout mechanism used by DNA binding
proteins.
Evolution
of Bacteriophage-encoded Exotoxins
Bacterially-derived exotoxins are among the most deadly
substances known. The genes that encode these exotoxins are usually carried by
bacterial viruses (bacteriophages) integrated into the bacterial host
chromosome. It is generally assumed that the targets of these toxins are
mammals. However, these phage-encoded exotoxin genes are widespread in the
environment and are found with unexpectedly high frequency in regions that lack
the presumed mammalian targets. These observations suggest that humans and other
susceptible mammals are not the primary “targets” of these toxins. We are
exploring the hypothesis that exotoxins are part of an antipredator defense
mechanism.
SELECTED PROJECTS
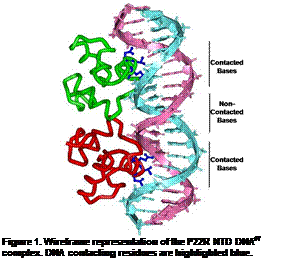 Indirect Readout: DNA Structure Effects on
Protein-DNA Interactions
Indirect Readout: DNA Structure Effects on
Protein-DNA Interactions
The binding of proteins to specific DNA
sequences plays a central role in the regulation of gene expression in all organisms.
These proteins regulate gene expression by binding DNA at specific sites and
activating or repressing transcription. Structural and biochemical studies have
provided a detailed insight into how the intimate contacts between proteins and
DNA enable proteins to bind specifically and with high affinity only to their cognate
DNA binding sites. One conclusion of these studies is that sequence specific
DNA recognition involves both direct and indirect readout of the binding site
sequence.
In indirect readout, the
stability and specificity of a protein-DNA complex is regulated by the sequence
of bases not in contact with the protein. These noncontacted bases can inhibit
or prevent the contacted DNA from being properly juxtaposed with protein
groups. DNA sequence-dependent differences in the structure and flexibility of
noncontacted bases lead to alterations in the strength and/or ease of forming
protein-DNA contacts. The DNA sequence-limited geometry changes thereby
indirectly alter the affinity and/or specificity of a protein for its cognate
binding site. Hence, indirect effects of DNA sequence on protein-DNA complex
formation occur by a modulation of the structural complementarity between the
interacting molecules.
While it is clear that
indirect readout of DNA sequence is an important component of the DNA
sequence recognition mechanisms of many proteins, until recently, it was
unclear how DNA sequence directs changes in DNA structure and/or flexibility.
Our current studies indicate that indirect readout by DNA binding proteins
repressor is influenced by sequence-dependent interactions between the solvent
and the unbound and/or protein-bound DNA. These data also show that
manipulating the solvent environment inside cells leads to specific effects on gene regulation.
Hence,
our ongoing studies are aimed at 1) providing a thermodynamic and structural
framework to explain the mechanisms of solvent-dependent indirect readout of
DNA sequence, and 2) understanding how these sequence- and solvent-dependent
differences in DNA structure influence the stability and function of
protein-DNA complexes.
 Evolution of Bacteriophage-encoded Exotoxins
Evolution of Bacteriophage-encoded Exotoxins
Phages encoding exotoxin genes are found ubiquitously as
lysogens in environmental bacteria, but it is unclear what advantage there is
to the bacteria to harbor phage that encode such toxic compounds. In the context of humans, these exotoxins cause diseases
ranging from cholera to diphtheria to enterohemorrhagic diarrhea. However, the
frequency of occurrence of the genes encoding any particular
exotoxin gene in bacteriophage and/or lysogens far exceeds the number of
potential animal hosts. Moreover, these phage-encoded exotoxin genes are found
at high frequency in free phages and lysogenic bacteria isolated from
environments where the corresponding human diseases are not prevalent. These
observations suggest that mammals are neither the original nor primary
“targets” of these toxins. The phage-encoded exotoxins like the well-studied
Shiga toxin (Stx), kill eukaryotic cells by attacking
features and pathways that are common to all eukaryotes both, uni- and multi-cellular. Thus the
evolution of these toxins may have occurred before the appearance of
multicellular organisms. Since predation by eukaryotic predators (e.g.,
ciliates and other protozoa), is a major source of bacterial mortality, these
observations suggest that exotoxins may have arisen as part of an antipredator,
(antiprotozoan) defense strategy. Hence humans may be
innocent bystanders in the evolutionary battle between protozoans and their
bacterial prey.
The environment in which microbes live is dynamic, changing as a consequence of anthropogenic, environmental and
evolutionary processes. Rapid changes can also result from the activities of
the microbes themselves when they respond to ecological pressures such as
predation. Our
published data indicate phage-encoded exotoxins (e.g., Shiga toxin, diphtheria
toxin) evolved as a defense against bacterivorous
predators. Our preliminary data indicate that the efficacy of an
exotoxin’s antipredator activity may govern the environmental persistence of
exotoxin-encoding bacteria and phages. By
determining how biochemical, cell biological and population-based factors
impact the persistence of an evolutionarily diverse set of Shiga toxin-encoding
bacteria and phage in natural and artificial microcosms, we are attempting to
1) identify how the microbial responses to predation shape, and are shaped by,
the microbial community and; 2) delineate how these responses impact microbial
survival and success.
SELECTED
PUBLICATIONS
(For complete list see here)
Gong, C,
Chakraborty D., Koudelka, G.B. (2023)
A prophage encoded ribosomal RNA methyltransferase regulates the
virulence of Shiga-toxin-producing Escherichia
coli (STEC) Nucleic Acids Res.
2023 Dec 12:gkad1150. doi:
10.1093/nar/gkad1150 https://doi.org/10.1093/nar/gkad1150.
Gong, C., Koudelka, G.B. (2023)
The Shiga toxin (Stx)-Phage Encoded Ribosomal RNA Methyltransferase
Regulates Stx-producing Escherichia coli (STEC) Virulence by Blocking Stx-Mediated
Inactivation of Bacterial Ribosomes (2023) bioRxiv.09.20.558713;
doi: https://doi.org/10.1101/2023.09.20.558713
Liu, Y., Koudelka, G.B. (2023)
O-polysaccharides of LPS modulate
E. coli uptake by Acanthamoeba. castellanii Microorganisms,
11, 1377 https://doi.org/10.3390/microorganisms11061377
.
Liu, Y., Koudelka, G.B. (2023)
The oligosaccharide region of LPS governs predation of E. coli by the bacterivorous
protist, Acanthamoeba castellanii Microbiology
Spectrum, 11 e02930-22 https://journals.asm.org/doi/10.1128/spectrum.02930-22
Peng, L.; Dumevi,
R.M.; Chitto, M.; Haarmann,
N.; Berger, P.; Koudelka, G.; Schmidt, H.; Mellmann,
A.; Dobrindt, U.; Berger, M. A (2022)
Robust One-Step Recombineering
System for Enterohemorrhagic Escherichia coli. Microorganisms, 10, 1689. https://doi.org/10.3390/microorganisms10091689
Lama, R., Xu, C., Galster, S.L., Querol-García, J., Portwood, S.,
Mavis, C.K., Ruiz, F.M., Martin, D., Wu, J., Giorgi, M.C., Bargonetti,
J., Wang, E.S., Hernandez-Ilizaliturri, F.J., Koudelka, G.B., Chemler,
S.R., Muñoz, Wang,
X. (2022)
Small Molecule MMRi62 Targets
MDM4 for Degradation and Induces Leukemic Cell Apoptosis Regardless of p53
Status Front. Oncol. https://doi.org/10.3389/fonc.2022.933446.
Berger, M., Aijaz, I., Berger, P., Dobrindt, U., Koudelka, G.B.
(2019)
Transcriptional and Translational
Inhibitors Block SOS Response and Shiga Toxin Expression in Enterohemorrhagic Escherichia coli.
Sci Rep 9, 18777. https://doi.org/10.1038/s41598-019-55332-2.
Berger, P., Kouzel, I.U., Berger, M., Haarmann, N., Dobrindt, U., Koudelka,
G.B., Mellmann, A. (2019)
Carriage of Shiga toxin phage
profoundly affects Escherichia coli
gene expression and carbon source utilization.
BMC Genomics 20:504.
doi.org/10.1186/s12864-019-5892-x.
Koudelka G.B., Arnold J.W.,
Chakraborty, D. (2018)
Evolution of STEC virulence: Insights from the
antipredator activities of Shiga toxin producing E. coli.
Int J Med Microbiol. 308 956-961. doi:
10.1016/j.ijmm.2018.07.001
Chakraborty, D., Clark, E.,
Mauro, S.A., Koudelka, G.B. (2018)
Molecular Mechanisms Governing
"Hair-Trigger" Induction of Shiga Toxin-Encoding Prophages.
Viruses 10 pii: E228. doi: 10.3390/v10050228.
Aijaz, I., Koudelka, G.B. (2018).
Cheating, facilitation and cooperation
regulate the effectiveness of phage-encoded exotoxins as antipredator molecules
Microbiologyopen e00636. doi: 10.1002/mbo3.636.
Aijaz, I., Koudelka, G.B. (2017)
Tetrahymena phagocytic
vesicles as ecological micro-niches of phage transfer
FEMS Microbiology Ecology, 93,
Issue 4, fix030, doi: 10.1093/femsec/fix030
Colon, M.P., Chakraborty D., Pevzner, Y., Koudelka. GB (2016)
Mechanisms determining the
differential stability of of Stx+
and Stx- lysogens
Toxins 8, 96
doi:10.3390/toxins8040096 (Full text).
Arnold, J.W.,
Spacht, D., Koudelka, G.B., (2016)
Determinants that govern the recognition and uptake of E. coli O157:H7 by Acanthamoeba castellanii
Cellular Microbiology, (2016) 18, 1459–1470 (Full text)
Samorodnitsky,
D., Szyjka, C., Koudelka, G.B., (2015)
A
Role for Autoinhibition in Preventing Dimerization of the Transcription Factor
ETS1
J.
Biol. Chem. 290, 22101-22110
(Full text)
Harris,
L.A., Williams, L.D., and Koudelka G. B. (2014)
Specific
minor groove solvation is a crucial determinant of DNA binding site
recognition.
Nucl.
Acids Res. doi:
10.1093/nar/gku1259 (Full text).
Arnold,
J.W, Koudelka, G.B. (2014)
Env.
Micro. 16, 454-466 (Full text).
Mauro,
S.A., Opalka, H., Lindsay, K., Colon, M.P., Koudelka, G.B. (2013)
Applied
and Env. Microbiology, 79, 4821-4842 (Full text).
Shkilnyj,
P., Colon, M.P., and Koudelka, G.B., (2013)
Bacteriophage
434 Hex protein prevents RecA-mediated repressor autocleavage
Viruses 5, 111-126 (Full text)
Stolfa,
G., and Koudelka, G.B., (2013)
Entry
and Killing of Tetrahymena by Bacterially Produced Shiga toxin,
mBio, 4 e00416-12; doi:10.1128/mBio.00416-12. (Full text)
Harris,
L.A., Watkins, D., Williams, L.D., and Koudelka G. B. (2013)
J.
Mol. Biol 425, 133-143., http://dx.doi.org/10.1016/j.jmb.2012.10.008.
Bullwinkle,
T.J., Samorodnitsky, D., Rosati, R.C. and Koudelka, G.B. (2012)
DNA
binding specificity determinants of 933W repressor,
PLOS
One, 7: e34563. doi:10.1371/journal.pone.0034563
Pawlowski
, D.R., Raslawsky, A., Siebert, G., Metzger, D.J., Koudelka, G.B., Karalus,
R.J. (2011)
Identification
of Hylemonella gracilis as an antagonist of Yersinia pestis persistence.
Journal
of Bioterrorism & Biodefense, S3:004. doi:10.4172/2157-2526.S3-004.
Mauro
S.A. and Koudelka, G.B (2011)
Shiga
Toxin: Expression, Distribution, and Its Role in the Environment
Toxins 3, 608-625; doi:10.3390/toxins3060608
Bullwinkle,
T.J., and Koudelka (2011)
J.
Bacteriol, 193, 3313-3323 (Full text)
Watkins,
D., Mohan, S., Koudelka, G.B., Williams, L.D., (2010)
Sequence
Recognition of DNA by Protein-Induced Conformational Transitions.
J.
Mol. Biol.
396, 1145-64. (Full text)
Lainhart,
W, Stolfa, G. and Koudelka, G.B. (2009)
Shiga
Toxin as a Bacterial Defense against a Eukaryotic Predator, Tetrahymena
thermophila,
J.
Bacteriol. 191
5116-5122 (Full text)
Page last
modified: 03/21/2024 by G. Koudelka
Return to the directory
Return to the Biological
Sciences Main
Page